Microbial Degradation of Nitrate: Put Microbes to Work DOI: 10.32526/ennrj.17.2.2019.10
Main Article Content
Abstract
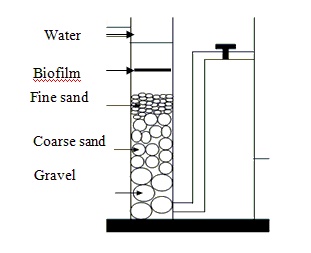
Three nitrate degrading bacteria, namely S1, S2 and S3 strains, were isolated from soil samples collected from agricultural sites at Polonnaruwa, Oruwala and Gampaha, Sri Lanka respectively. Among the isolated strains, S1 showed a maximum nitrate removal rate of 4.20±0.08 mg/L/day whereas S2 and S3 showed nitrate removal rates of 3.45±0.57 mg/L/day and 3.72±0.19 mg/L/day, respectively. The nitrate removal abilities of all three strains were measured at different temperatures (25oC, 30oC, 32oC), different pH (7.0, 7.5, 8.0) and for different nitrate concentrations (15, 30, 45 mg/L) in vitro. The maximum nitrate degrading rate by the bacterium S1 (4.52±0.01 mg/L/day), S2 (4.25±0.47 mg/L/day) and S3 (4.04±0.09 mg/L/day) were detected at 32ºC whereas the highest degradation rate for S1 (4.6±0.05 mg/L/day), S2 (4.38±0.03 mg/L/day) and S3 (4.14±0.25 mg/L/day) bacteria strains was reported when the pH was 8.0. The maximum nitrate degradation was found to be 5.88±0.17, 5.67±0.45 and 5.71±0.43 mg/L/day for S1, S2 and S3 bacteria strains when 45 mg/L of nitrate was present in the medium. A biochemical test tentatively identified the isolates S1, S2 and S3 as Pseudomonas sp., Bacillus sp. and Proteus sp. The bio sand filter developed in co-operating all three bacterial strains showed 6.12±0.06 mg/L/day of nitrate degradation rate within 24 hours.
Article Details
Published articles are under the copyright of the Environment and Natural Resources Journal effective when the article is accepted for publication thus granting Environment and Natural Resources Journal all rights for the work so that both parties may be protected from the consequences of unauthorized use. Partially or totally publication of an article elsewhere is possible only after the consent from the editors.
References
2. Aopreeya M, Arunlertaree C, Yuwaree C, Boonprasert R, Hutacharoen R. Blood cockle shell: an agro-waste for N and P removal of shrimp farm effluent. Environment and Natural Resources Journal 2013;11(1):58-69.
3. Bonse U, Busch F, Günnewig O, Beckmann F, Pahl R, Delling G, Hahn M, Graeff W. 3D computed X-ray tomography of human cancellous bone at 8 μm spatial and 10-4 energy resolution. Bone and Mineral 1994;25(1):25-38.
4. Burri R, Stutzer A. Ueber Nitrat zerstörende Bakterien und den durch dieselben bedingten Stickstoffverlust. Zentbl Bakteriol Parasitenkd 1895;Abt.II(1):257-65.
5. Cabello P, Roldan MD, Moreno-Vivian C. Nitrate reduction and the nitrogen cycle in archaea. Microbiology 2004;150(11):3527-46.
6. Cambra-López M, Aarnink AJ, Zhao Y, Calvet S, Torres AG. Airborne particulate matter from livestock production systems: a review of an air pollution problem. Environmental pollution 2010;158(1):1-17.
7. Carlson CA, Ingraham JL. Comparison of denitrification by Pseudomnas stutzeri, Pseuodomonas aeruginosa and Paracoccus dentrificans. Applied and Environmental Microbiology 1983;45(4):1247-53.
8. Choi JW, Wicker R, Lee SH, Choi KH, Ha CS, Chung I. Fabrication of 3D biocompatible/biodegradable micro-scaffolds using dynamic mask projection microstereolithography. Journal of Materials Processing Technology 2009;209(15-16):5494-03.
9. Cowan ST, Steel KJ. Cowan and Steel's Manual for the Identification of Medical Bacteria. Cambridge University Press; 2004.
10. DebRoy S, Mukherjee P, Roy S, Thakur AR, RayChaudhuri S. Draft genome sequence of nitrate- and phosphate-removing Bacillus sp., WBUNB009. Genome Announcements 2013;1(1):251-63.
11. Della Valle CJ, Scher DM, Kim YH, Oxley CM, Desai P, Zuckerman JD, Di Cesare PE. The role of intraoperative Gram stain in revision total joint arthroplasty. Journal of Arthroplasty 1999;14(4):500-4.
12. Eckford RE, Fedorak PM. Planktonic nitrate-reducing bacteria and sulfate-reducing bacteria in some western Canadian oil field waters. Journal of Industrial Microbiology and Biotechnology 2002;29(2):83-92.
13. Ergas SJ, Reuss, AF. Hydrogenotrophic denitrification of drinking water using a hollow fibre membrane bioreactor. Journal of Water Supply: Research and Technology-Aqua 2001;50(3):161-71.
14. Gao W, Jin R, Chen J, Guan X, Zeng H, Zhang F, Guan N. Titania-supported bimetallic catalysts for photocatalytic reduction of nitrate. Catalysis Today 2004;90(3-4):331-6.
15. Gómez MA, Hontoria E, González-López J. Effect of dissolved oxygen concentration on nitrate removal from groundwater using a denitrifying submerged filter. Journal of Hazardous Materials 2002:90(3):267-78.
16. Hettiarachchi IU, Sethunga S, Manage PM. Contamination Status of Algae Toxins Microcystins in Some Selected Water Bodies in Sri Lanka. Proceedings of the International Forestry and Environment Symposium; 2014 Jan 11; MAS Economic Zone, Sri Lanka; 2014.
17. Idroos FS, Manage PM. Seasonal occurrence of MIcrocystin-LR with respect to physico- chemical aspects of Beira lake water. International Journal of Multidisciplinary Studies 2014;1(2):27-37.
18. Idroos FS, De Silva BGDNK, Manage PM. Biodegradation of microcystin analogues by Stenotrophomonas maltophilia isolated from Beira Lake Sri Lanka. Journal of the National Science Foundation of Sri Lanka 2017;45(2):91-9.
19. Janda JM, Abbott SL. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations, and unanswered questions. Clinical Infectious Diseases 1998;27(2):332-44.
20. Koenig A, Liu LH. Autotrophic denitrification of landfill leachate using elemental sulphur. Water Science and Technology 1996;34(5-6):469-76.
21. Lampe DG, Zhang TC. Evaluation of Sulfur-based Autotrophic Denitrification [dissertation]. Lincoln: University of Nebraska; 1996.
22. Lim YW, Lee SA, Kim SB, Yong HY, Yeon SH, Park YK, Jeong DW, Park JS. Diversity of denitrifying bacteria isolated from Daejeon sewage treatment plant. Journal of Microbiology 2005;43(5):383-90.
23. Mahagamage MGY, Pathmalal M. Water Quality Index (CCME-WQI) Based Assessment Study of Water Quality in Kelani River Basin, Sri Lanka. Proceedings of the International Environment and Natural Resources Conference; 2014 Nov 6-7; The Sukosol Hotel, Bangkok: Thailand; 2014.
24. Manage PM, Edwards C, Singh BK, Lawton LA. Isolation and identification of novel microcystin-degrading bacteria. Applied and Environmental Microbiology 2009;75(21):6924-8.
25. Mishra CN, Tiwari V, Satish-kumar VG, Kumar A, Sharma I. Genetic diversity and genotype by trait analysis for agromorphological and physiological traits of wheat (Triticum aestivum L.). Sabrao Journal of Breeding and Genetics 2015;47(1):40-8.
26. Mohammadi AS, Movahedian H, Nikaeen M. Drinking water denitrification with autotrophic denitrifying bacteria in a fluidized bed bioreactor (FBBR). Fresenius Environmental Bulletin 2011;20(9A):2427-36.
27. Mohseni-Bandpi A, Elliott DJ, Zazouli MA. Biological nitrate removal processes from drinking water supply: a review. Journal of Environmental Health Science and Engineering 2013;11(1):35.
28. Mousavi SAR, Ibrahim, S, Aroua MK, Ghafari, S. Bio-electrochemical denitrification: a review. International Journal of Chemistry and Environmental Engineering 2011;2(2).
29. Percheron G, Bernet N, Moletta R. Interactions between methanogenic and nitrate reducing bacteria during the anaerobic digestion of an industrial sulfate rich wastewater. FEMS Microbiology Ecology 1999; 29(4):341-50.
30. Rajkumar M, Nagendran R, Lee KJ, Lee WH, Kim SZ. Influence of plant growth promoting bacteria and Cr6+ on the growth of Indian mustard. Chemosphere 2006;62(5):741-8.
31. Rezaei F, Xing D, Wagner R, Regan JM, Richard TL, Logan BE. Simultaneous cellulose degradation and electricity production by Enterobacter cloacae in a microbial fuel cell. Applied and Environmental Microbiology 2009;75(11):3673-8.
32. Sahinkaya E, Kilic A, Duygulu B. Pilot and full scale applications of sulfur-based autotrophic denitrification process for nitrate removal from activated sludge process effluent. Water Research 2014;60:210-7.
33. Shrimali M, Singh KP. New methods of nitrate removal from water. Environmental Pollution 2001;112(3): 351-9.
34. Sri Lanka Standards Institute (SLSI). Drinking Water Guidelines. SLS 614: 2013.
35. Su HL, Chou CC, Hung DJ, Lin SH, Pao IC, Lin JH, Huang FL, Dong RX, Lin JJ. The disruption of bacterial membrane integrity through ROS generation induced by nanohybrids of silver and clay. Biomaterials 2009;30(30):5979-87.
36. Wakida FT, Lerner DN. Potential nitrate leaching to groundwater from house building. Hydrological Processes 2006;20(9):2077-81.
37. Wang Q, Feng C, Zhao Y, Hao C. Denitrification of nitrate contaminated groundwater with a fiber-based biofilm reactor. Bioresource Technology 2009;100(7): 2223-7.
38. Wenske H, Matschiner H, Siegel H. Electrolytic cell and capillary gap electrode for gas-developing or gas-consuming electrolytic reactions and electrolysis process therefor. Maschinen- und Anlagenbau Grimma GmbH MAG. 1997.
39. Wick K, Heumesser C, Schmid E. Groundwater nitrate contamination: factors and indicators. Journal of Environmental Management 2012;111:178-86.
41. Yang WS, Park BW, Jung EH, Jeon NJ, Kim YC, Lee DU, Shin SS, Seo J, Kim EK, Noh JH, Seok SI. Iodide management in formamidinium-lead-halide–based perovskite layers for efficient solar cells. Science 2017;356(6345):1376-9.
42. Zumft WG. Cell biology and molecular basis of denitrification. Microbiology and Molecular Biology Reviews 1997;61(4):533-616.